Views Types
Vitessce supports several view types for visualization of single-cell data, and several view types for updating visualization parameters. The view names found on this page are all valid values for the field layout[].component in the view config. On this page you will also find screenshots of each view.
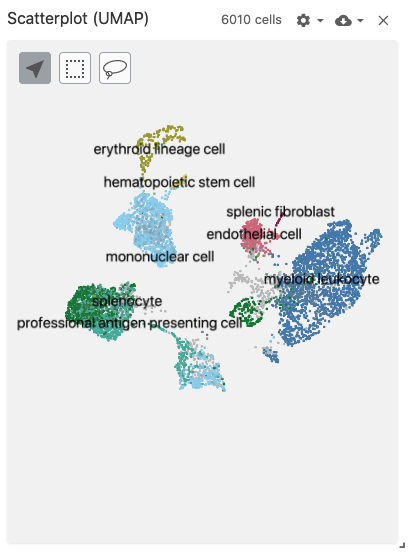
scatterplot
The scatterplot view displays 2-dimensional (pre-computed) embeddings / projections (such as t-SNE or UMAP).
Use with the embeddingType coordination type to specify which embedding to use when mapping cells onto scatterplot points.
Use with datasets containing the obsEmbedding data type. Optionally, the obsSets and obsFeatureMatrix data types can be used for coloring cells on the plot.
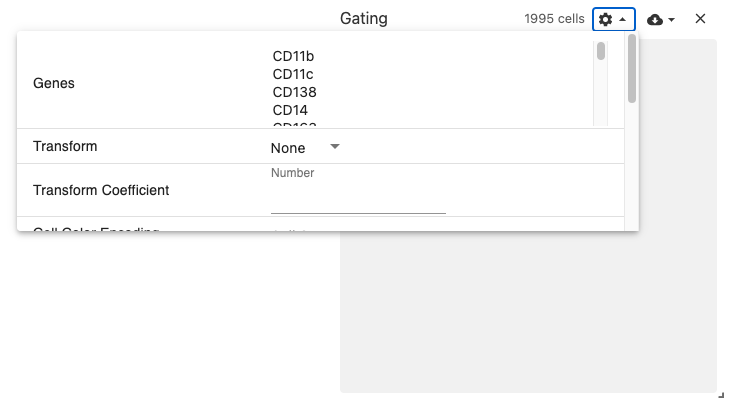
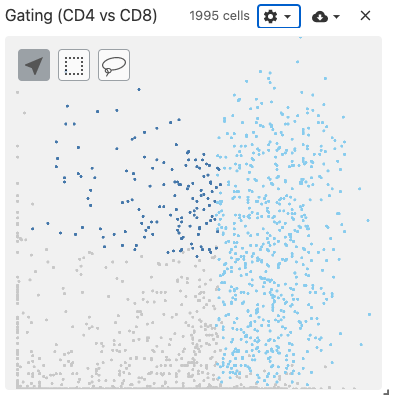
gating
The gating view displays 2-dimensional scatterplot of gene expression data.
Users select two genes, and a scatterplot is dynamically generated using the obsFeatureMatrix data. Gating can then be performed by using the lasso or box select tools.
Use with datasets containing the obsFeatureMatrix data types. Optionally, the obsSets and obsFeatureMatrix data types can be used for coloring cells on the plot.
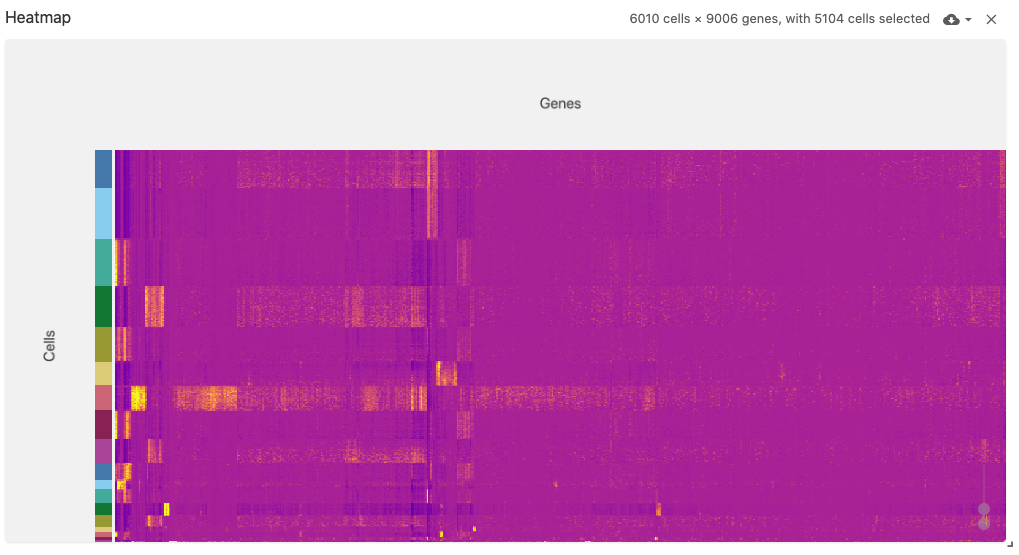
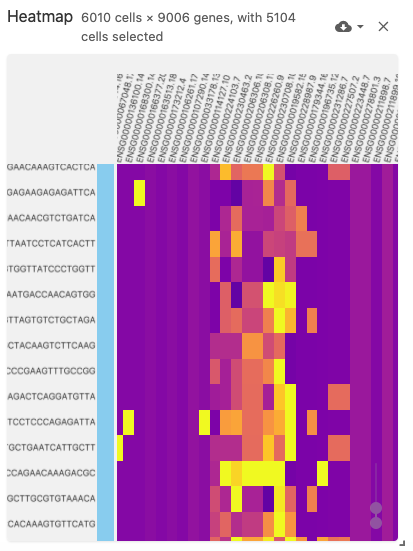
heatmap
The heatmap view displays a (normalized) cell-by-gene (or gene-by-cell) matrix visualization.
This view uses the obsFeatureMatrix data type, and optionally displays cell set color assignments if a file with the obsSets data type is available in the same dataset.
The heatmap can support very large matrices (~6,000 x ~9,000) when using Zarr-based file types (best performance can be acheived when arrays are saved with the type uint8).
spatial
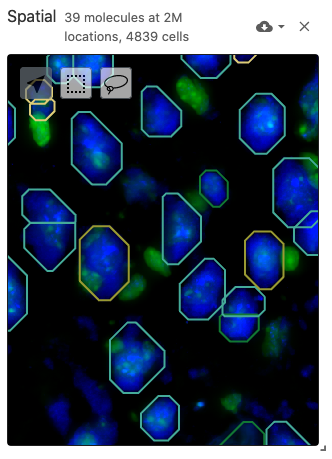
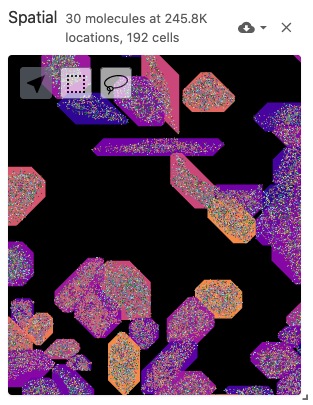
The spatial view is meant to display data with spatial coordinates, including spatially-resolved cell segmentations as polygons (from the obsSegmentations data type) and molecule positions as points (from the obsLocations data type). The spatial view also includes a multiplexed and multi-scale image viewer (implemented with Viv). Image files can be defined with the image data type. Cell segmentations can be colored by gene expression data through the obsFeatureMatrix data type or cell set color assignments through the obsSets data type.
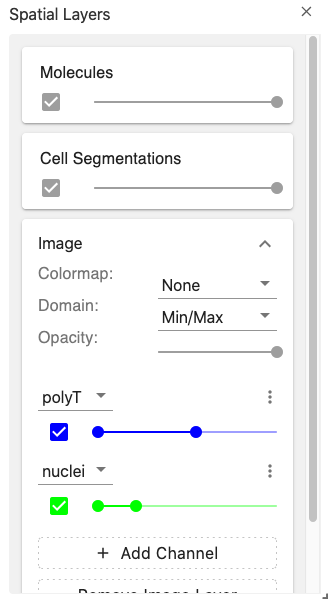
layerController
The layer controller view provides an interface for manipulating the visualization layers displayed in the spatial view.
The image layers are coordinated through the spatialImageLayer coordination type.
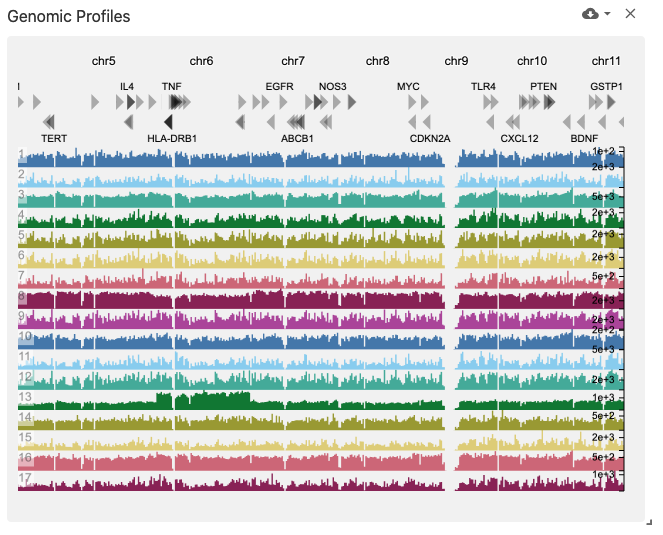
genomicProfiles
The genomic profiles view displays genome browser tracks (using the genomic-profiles data type) containing bar plots, where the genome is along the x-axis and the value at each genome position is encoded with a bar along the y-axis.
Genome tracks may be colored by corresponding cell set color assignments if the obsSets data type is available in the same dataset, coordinated on the obsSetSelection coordination type.
This view was implemented with HiGlass.
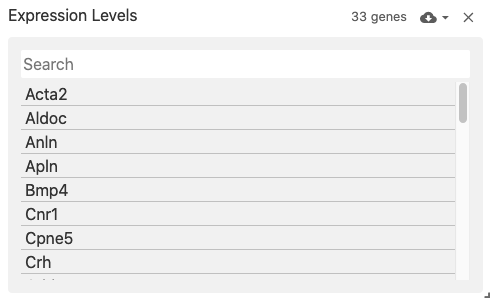
featureList
The feature list view displays an interactive list of features (i.e., genes when featureType: 'gene').
The list is obtained from the feature axis of matrix files corresponding to the obsFeatureMatrix data type.
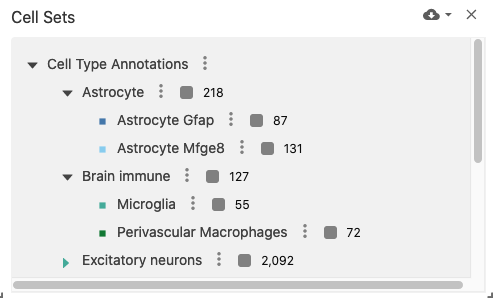
obsSets
The observation sets view displays an interactive list of (potentially hierarchical) observation sets (e.g., cell sets) when used with datasets containing the obsSets data type. This can be useful for managing cell sets representing clustering algorithm outputs or cell type annotations.
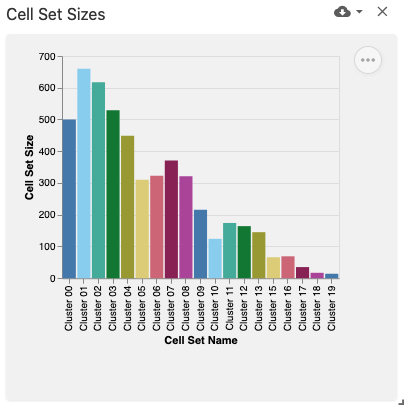
obsSetSizes
The observation set sizes view displays a bar plot with the currently-selected observation sets (e.g., cell sets) using the obsSets data type and obsSetSelection coordination type on the x-axis and bars representing their size (e.g., by number of cells) on the y-axis. This view was implemented with Vega-Lite.
description
The description view can be used to display text content. This may be useful for communicating the details of a dataset or a data analysis process. When the spatial view is used for visualization of imaging data, the description view also renders a dropdown containing image metadata.

status
The status view displays debugging messages, including app-wide error messages when datasets fail to load or when schemas fail to validate. Details about the entity under the mouse cursor (cell, gene, and/or molecule) are displayed during hover interactions.

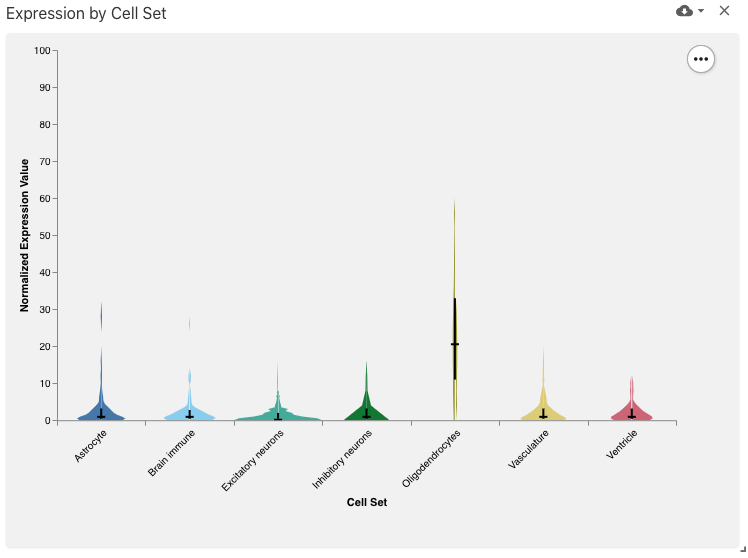
obsSetFeatureValueDistribution
The observation set feature value distribution view displays a violin plot with values (e.g., expression values) for the selected feature (e.g., gene), implemented with Vega-Lite.
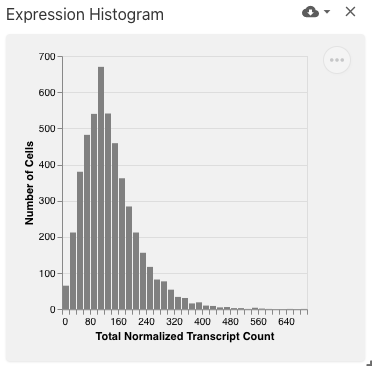
featureValueHistogram
The feature value histogram view displays the distribution of values (e.g., expression) for the selected feature (e.g., gene), implemented with Vega-Lite.